Group of Dr. Traube - Faculty for Chemistry and Pharmacy
Group of Dr. Traube - Faculty for Chemistry and Pharmacy
 Group of Dr. Traube - Faculty for Chemistry and Pharmacy
Group of Dr. Traube - Faculty for Chemistry and Pharmacy
Our group investigates how epigenetic markers are placed and removed during development and disease. We are especially interested in how metabolic decisions influence epigenetic patterns (Research Area A), how epigenetic dysregulation and proliferative signaling affect each other during tumorigenesis (Research Area B) and how epigenetic aberrations that promote tumorigenesis can be permanently changed (Research Area C).
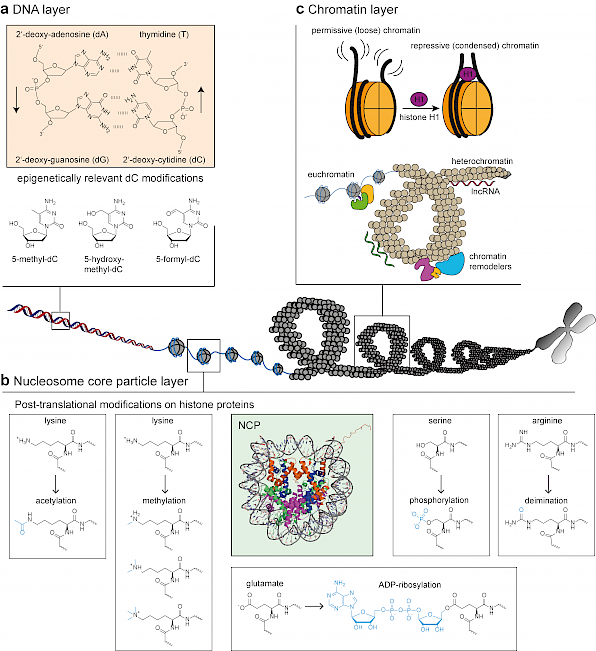
To develop and function correctly, cells of multicellular organisms rely on inheritable yet dynamic epigenetic changes that enable cell-type specific gene expression patterns, but do not change the underlying genetic information. Epigenetic regulation occurs on the DNA (a) and RNA level in form of nucleobase modifications, on the nucleosome level in form of post-translational histone modifications (b) and on the global chromatin level by changing chromatin accessibility (c). The most dramatic changes of the epigenetic landscape happen during differentiation when pluripotent stem cells undergo lineage commitment to become all sorts of specialized cells. Epigenetic aberrations during this process often result in cancer development as the affected cells do not undergo the epigenetically introduced switch from proliferation to maturation with the associated changes in gene expression anymore.