 Group of Dr. Traube - Faculty for Chemistry and Pharmacy
Group of Dr. Traube - Faculty for Chemistry and Pharmacy
 Group of Dr. Traube - Faculty for Chemistry and Pharmacy
Group of Dr. Traube - Faculty for Chemistry and Pharmacy
In recent years, the metabolic control of the epigenetic landscape has moved into the spotlight of epigenetic research, since metabolites like α-ketoglutarate (αKG), acetyl-CoA or S-adenosyl-methionine (SAM) determine the activity of epigenetic enzymes. Especially fluctuations of αKG, an essential co-substrate for ten-eleven translocation (TET) enzymes, which oxidize 5-methyl-2'-deoxy-cytidine, and for Jumonji-C domain containing histone demethylases (JHDMs), turned out to play a pivotal role to control epigenetic dynamics and in consequence differentiation.
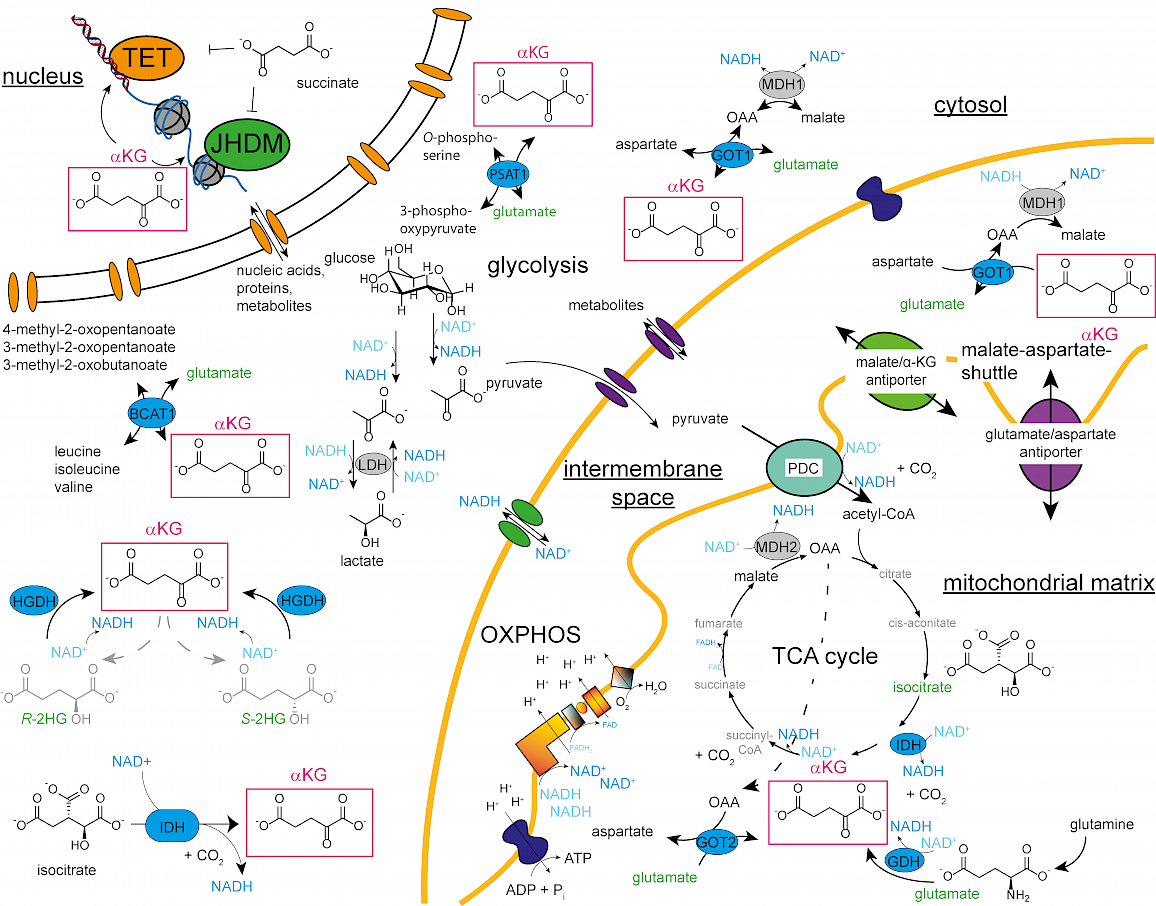
By modulating the availability of epigenetically relevant metabolites, the activity of epigenetic enzymes can be immediately enhanced, slowed down or even stopped without changing overall protein levels that need much longer to react to extra- and intracellular stimuli. Following a holistic approach, we apply a wide range of methods from (molecular) cell biology and analytical chemistry to uncover how metabolic decisions influence epigenetic patterns and vice versa in differentiating cells during hematopoiesis and neurogenesis. We combine information from LC-MS/MS and imaging based proteome and metabolome profiling experiments to reveal how epigenetic enzymes are supplied with their required co-substrates. By changing the abundance, location or the activity of the metabolic supplier enzymes, we finally want to selectively interfere with this epigenetic/metabolic interplay during differentiation to understand on a molecular level the metabolic pre-requisites that underly epigenetic dynamics.
Topic-related publications:
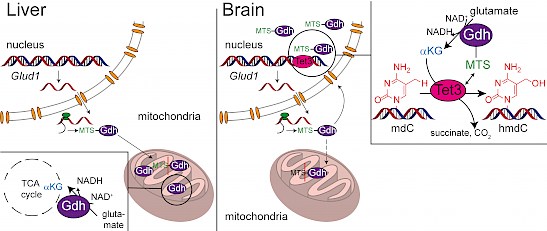
Traube FR, Özdemir D, Sahin H, Scheel C, Glück AF, Geserich AS, Oganesian S, Kostidis S, Iwan K, Rahimoff R, Giorgio G, Müller M, Spada F, Biel M, Cox J, Giera M, Michalakis S#, Carell T#. “Redirected nuclear glutamate dehydrogenase supplies Tet3 with a-ketoglutarate in neurons“ Nat. Commun. 12 (2021), 4100.
We discovered that neuronal Tet3, which is the most important TET enzyme in brain, requires on-site biosynthesis of αKG in the nucleus, which is provided by glutamate dehydrogenase (Gdh). Gdh has an N-terminal mitochondrial targeting sequence (MTS) and is therefore normally located to the mitochondria, where the MTS is cleaved off and the enzyme feed αKG from glutamate into the TCA cycle. In neurons, however, Gdh is redirected to the nucleus by Tet3 to supply Tet3 with αKG instead.
Traube FR*, Schiffers S*, Carell T#. “Quantification of DNA Methylation and Its Oxidized Derivatives Using LC-MS” Methods Mol. Biol. 2272 (2021), 77 – 94.
Traube FR*, Schiffers S*, Iwan K*, Kellner S, Spada F, Müller M, Carell T#. “Isotope-dilution mass spectrometry for exact quantification of noncanonical DNA nucleosides” Nat. Protoc. 14 (2019), 283 – 312.